

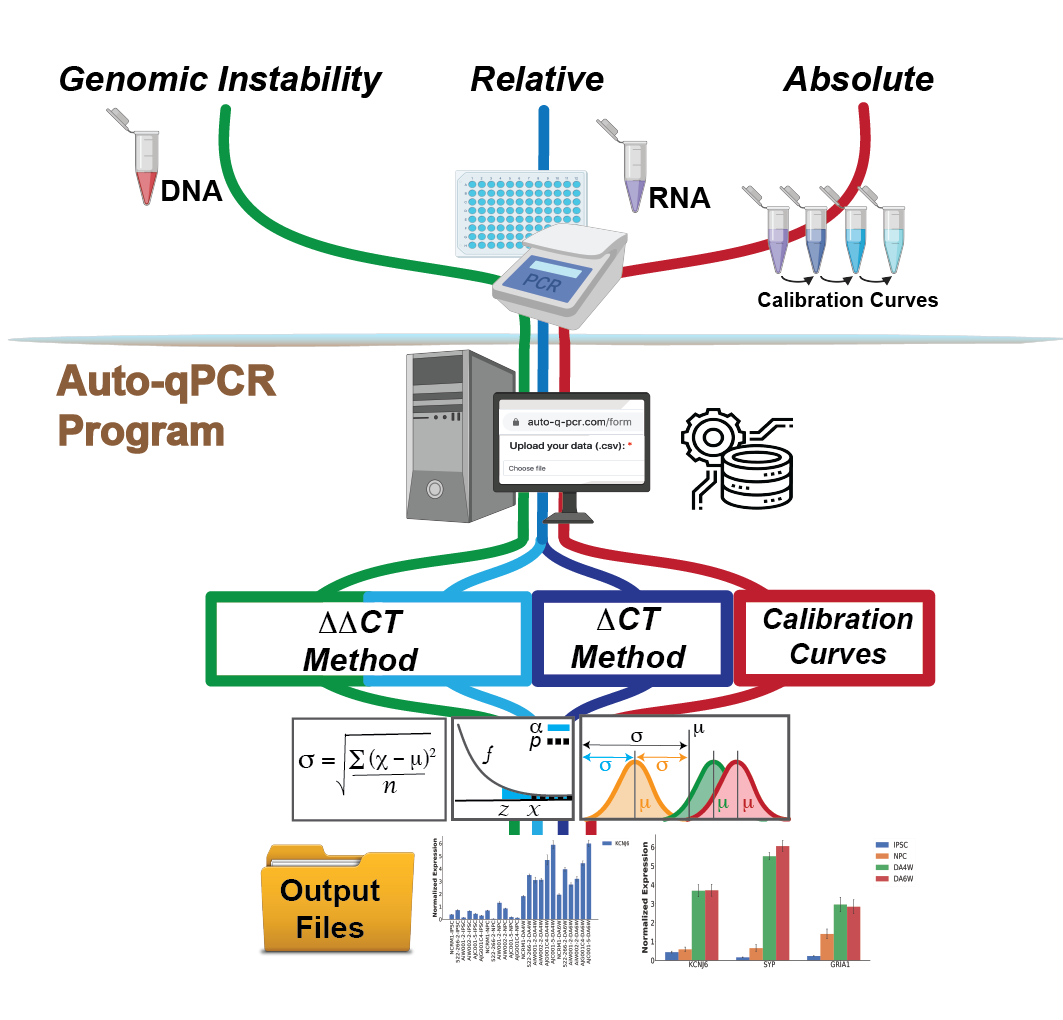
Auto-qPCR is a python based web app designed to process raw data files from thermal cycler machines.
Please cite the following manuscript if you use this program in your research:
Maussion, G.*, Thomas, R.A.*, Demirova, I. et al. Auto-qPCR; a python-based web app for automated and reproducible analysis of qPCR data. Sci Rep 11, 21293 (2021).*Equal contribution
https://doi.org/10.1038/s41598-021-99727-6
The source code for the program can be found at:
https://github.com/neuroeddu/Auto-qPCR
Quantifying changes at the DNA and RNA levels are essential components of any molecular biology toolkit and can be done using quantitative PCR technology.However, the analysis of qPCR data includes many steps that are time consuming and cumbersome and prone mistakes. We developed Auto-qPCR to address this bottleneck. Examples data for four the different modes of data processing can be found in the help tab, along with a downloadable version. Auto-qPCR has functions for the following types of analysis:
1) Absolute model of RNA quantification using a calibration curve
2) Relative quantification model
2) relative to endogenous control(s) (delta-CT)
3) relative to endogenous control(s) and a reference sample
4) cDNA quantification to identify deletion or duplication events within the genome using a target
reference and a reference sample (delta-delta-CT)
Copyright ©2019-2021. NeuroEDDU Rhalena Thomas, Gilles Maussion, Eddie Cai, Gracia Gu, Iveta Demirova and Thomas Durcan. This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation version 3 of the License, This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. .